The genetic genealogy testing companies were out in force at the i4GG conference this past weekend in San Diego! Representatives from Living DNA and MyHeritage gave hour-long talks on Saturday, and people from Family Tree DNA, AncestryDNA, and 23andMe spoke on Sunday.
They all gave polished and informative presentations, and I made a point to see them all so that I could report back to my readers. This is the fifth in a series of five (in the order of the original presentations) on what they had to say.
 Tracing Your DNA Across the Ages with 23andMe
Tracing Your DNA Across the Ages with 23andMe
Hilary Vance, Product Supervisor at 23andMe
The final company presentation at i4GG was by Hilary Vance of 23andMe. Similar to the AncestryDNA talk, this one was more an overview of the existing features at 23andMe than a sneak-peek of what’s to come.
At present, 77% of the 23andMe database is of European descent, and 85% consent to participate in the research program. My back-of-the-envelope calculations suggest that they have more than half a million testers of non-European descent who are contributing to scientific discovery. (One of my main concerns with biomedical research is that it’s too heavily focused on Europeans, so this number makes me happy.) Customers who have consented to research can go to the Research tab in their accounts, then Publications to see how many published papers have benefitted from their participation.
Ancient Migrations
Mitochondrial DNA (mtDNA) traces your matrilineal descent from a single woman known as “Mitochondrial Eve” who lived more than 150,000 years ago. All humans carry mtDNA descended from hers. (She wasn’t the only human woman living at the time, of course, nor the only one who left descendants, but the other women’s descendant lineages either failed to reproduce or “maled out”, with no daughters to pass on the mtDNA.) 23andMe‘s test samples some of the DNA In your mitochondria and uses it to assign you to a mitochondrial haplogroup. That report was updated in May 2017 and now includes more than 200 “origin” stories for various subbranches of the mitochondrial tree.
The figure below shows a mtDNA haplogroup migration map from the 23andMe blog dated 1 June 2017.
Analogous to Mitochondrial Eve was a man called Y-chromosomal Adam, who lived more than 275,000 years ago. (He was not a contemporary of Mitochondrial Eve.) The Y-chromosome haplogroup report at 23andMe is based on more than 2000 yDNA SNPs. It was updated in November 2016 to agree with the ISOGG haplotree and had a minor update in May 2017. It’s been changed to use shorthand names. (For example, my father’s designation changed from R1b1b2a1a2f2 to R-M222.) A scientist at 23andMe also developed yHaplo, an open-source program for non-commercial use to identify male haplogroups.
Women don’t have y chromosomes, but female customers can “share” their DNA results with a father or paternal brother to get yDNA reports themselves.
The final “ancient” report at 23andMe is Neanderthal Ancestry, showing how much of your genome can be attributed to our Homo sapiens ancestors mating with Neanderthals sometime between 60,000 and 40,000 years ago. Most 23andMe customers have 200–400 Neanderthal variants (in 600,000 or so total SNPs).
Ancestry Composition
The “ethnicity estimates” at 23andMe are in the Ancestry Composition report, which aims to reflect back about 500 years. Like such reports at the other companies, it is based on a set of reference individuals, who are unrelated people with deep roots in each specific area. The reference populations were updated when 23andMe switched to their latest chip (version 5).
23andMe maps the regional estimates onto your chromosomes, so you can see which sections of your genome are likely inherited from which geographic areas. To help separate the markers on your maternally inherited chromosomes from your paternally inherited ones, 23andMe uses a program called “Eagle” to statistically phase our data for the ethnicity estimates. They do an initial pass over the chromosomes to assign populations, then a second draft corrects for potential errors. The end results is not a yes-or-no assignment but rather a probability that a given section of DNA came from each reference population.
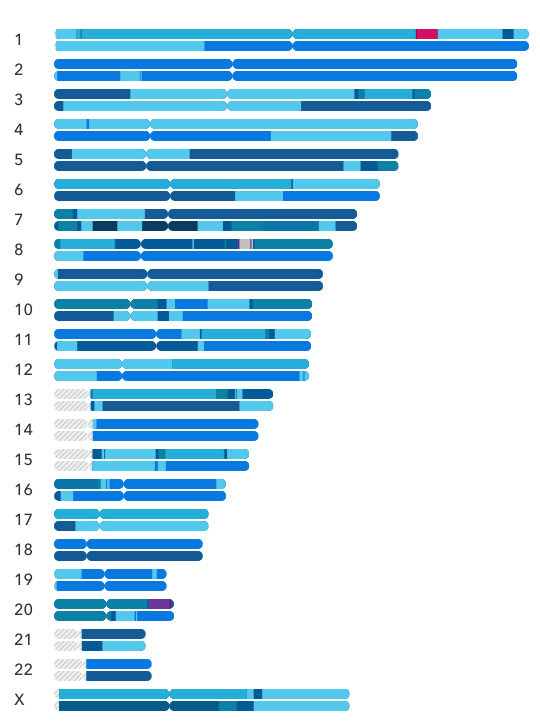
My Chromosome Painting is shown below. The shades of blue are various flavors of European, the hot pink on chromosome 1 is West African, and the dark purple on chromosome 20 is North African.
Other Features
In addition to the reports described above, 23andMe has a GrandTree tool that lets you explore how traits were inherited from your grandparents (if they have also tested at 23andMe ). Ms Vance also described the Traits Reports, which describe how your genetics affect your physical appearance, such as earlobe attachment or hair color.
Recent updates to the DNA Relatives feature include a filter for birth locations and the display of fully identical and half identical regions of DNA (both introduced March 2017).
The one potential “down the pike” tidbit was in response to a question from the audience: 23andMe is considering using statistical phasing for relative matching. This would give us greater confidence in small segment matches.
Other Posts in this Series
You can see what the other companies had to say at i4GG by following these links:




What is statistical phasing, and how would it improve confidence on small matches?
Small “matching” segments are frequently false positives caused by strand switching. For example, your maternal and paternal chromosomes may read:
AGATTC
ACAATC
but the technology does not allow us to tell which SNPs are on the same strand as which other SNPs. Together, your two chromosomes might appear to match a segment that reads:
AGAATC
even though neither of your parents gave you that sequence. That would be a false positive.
The smaller the segment, the more likely it is to be a false positive, with some estimates saying that a 7 cM segment has a 50-50 chance of being IBD versus a false positive.
With true phasing, you would use data from one or both of your parents to help sort out which SNPs belong together. But we can’t all genotype our parents. Statistical phasing uses knowledge of the haplotypes in a population to make educated guesses about which SNPs are on the same strand as which others. The “pseudophased” data will have fewer false positives than unphased data, and when you do have small matching segments, you can be more confident that they are real IBD.
Sounds like it will also eliminate and have fewer valid matches than unphased data.
You can read more about statistical phasing here:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2790566/pdf/main.pdf
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3217888/pdf/nihms326528.pdf
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3664855/pdf/459.pdf
Hi,
I am trying to contact dna land as I made a mistake on one of the surveys. I don’t use facebook. Is there an email for dna land so that I may correct my error?
Thank you very much,
Kim
They don’t seem to have contact information other than via Facebook.
https://dna.land/contact
Thank you. I am not going to do facebook though. I guess the mistake will have to be left. It is only on the height. I put 5′ 1 1/2″, and it put 170cm. I guess I should have put 5’2″. Obviously, that is an error. Thanks for trying to help. I wish they had a direct email.
Kim