This is my 100th post!
A special thanks to all of my readers for making blogging so much fun.
Scroll down for links to other posts in this series.
The first three posts in this series explained the underlying principles behind the probability approach to genetic genealogy: form two or more hypotheses about how an “unknown” person (or target) fits into a known tree, calculate how likely each hypothesis is, then focus on the most likely hypotheses for additional testing and/or paper-trail confirmation. The next three posts walked us through specific examples in which the method was applied. Those examples used a calculator in a table format; you enter a matrix of names, shared cM amounts, and hypothesized relationships to perform the calculations.
Tables, however, aren’t very intuitive when we’re dealing with branching trees, and it’s very easy to make an error when determining or entering the relationships. I often forget to specify half relationships, and typos are always a concern.
For the past few months, Jonny Perl of DNA Painter and I have been working on a much more intuitive way to test hypotheses. The math is the same, but the interface allows you to build a visual descendant tree rather than deal with a messy (and boring) table. Plus, it calculates the relationships for you!
Announcing … the What Are the Odds? tool! (WATO for short.)
What I love most about Jonny Perl’s programming is that he makes complex concepts intuitive and even fun. (Check out his DNA Painter chromosome mapper, if you haven’t already.) For that reason, I’m tempted to unleash you on WATO with no instructions and let you figure it out on your own. If that’s your cup of tea, have at it! If not, continue reading for a quick tutorial.
Before You Start
The goal of WATO is to help you figure out where an unknown person fits into a known tree. We’ll call that person the “target”. If you are an adoptee looking looking for birth family, you’re the target. Alternately, if you have a well-supported tree and an unknown match, they are the target, and this approach can help you figure out how they are related to your family.
Before you use the tool, your target person should have:
- Multiple DNA matches of 40 cM or more who are all descended in known ways from the same ancestor or couple (A few matches below 40 cM are fine, too)
- A descendant tree of the ancestral person/couple that includes the known DNA matches in their proper places
- The amount of shared DNA (cM are preferable, but percent will work) between the target person and each of those DNA matches. (The shared DNA amounts can come from any of the companies, but for FTDNA data, you must subtract out segments smaller than 7 cM first.)
- Two or more educated guesses about where the target person might fit into the tree
How to Use the Tool
To get started, go to this URL: https://dnapainter.com/tools/probability
You will be greeted with a set of instructions. Read through them if you like, then click “CLOSE INSTRUCTIONS” at the top right to dismiss them. The blank tool will look like this:
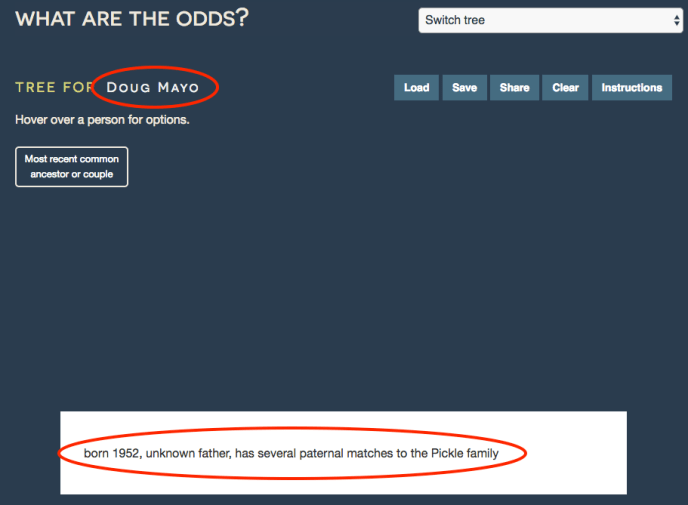
First, click on the text that says “Enter target name here” to title the tree. Then, enter a few notes to summarize the question at hand. This fictional example will try to identify the birth father of “Doug Mayo”, who has several DNA matches to the Pickle family.
Click on the box labeled “Most recent common ancestor or couple” and enter the name(s) of the common ancestor/couple of the DNA matches.
Hover your cursor over the box and click “Add child” in the menu option that appears. Repeat for each child of that couple who has a DNA-tested descendant, plus one. In our example, Peter Pickle and his wife Gladys Honey had seven children, but only four are represented by DNA tested descendants. I’ll add four children, plus a fifth to represent their other three children in our hypotheses.
Click on each child in turn to enter a name. Because no known descendants of Maisie, Gerald, or James Pickle have tested yet, I’ll lump them together in the same box for now to avoid cluttering the diagram.
Use the same approach to fill in the descendant tree, tracing down to their DNA-tested descendants. You don’t need to add every single descendant—just the ones that lead to DNA matches—and you don’t need know every person’s real name.
When you reach someone who is a DNA match to the target person, hover over their box, click “Enter Match cM”, enter the shared amount, and click “Save”. There’s an option to enter % shared if you prefer.
The box will change colors and show the cM amount.
Continue until all of the known DNA matches in the family have been accounted for. At this stage, my diagram looks like this:
Now we add “dummy” people where we think the target person might fit into the Pickle tree. It’s possible that Doug Mayo is descended from Maisie, Gerald, or James Pickle. I’m not sure which generation he’s in, so I’ll add three for now.
![]()
In each spot where Doug might fit, I hover over the dummy person and select “Use as Hypothesis”.
The hypothesis will be automatically numbered and assigned a “score” based on how likely it is relative to the other hypotheses. If there’s only one hypothesis, the score will always be either “1” (possible) or “0” (not possible). Because the scores are based on comparisons to the other hypotheses and are not absolute scores, they will change as I add more hypotheses.
I think it’s also possible that Doug is descended from Jasper Pickle, either through a full sibling to JJ and Annie or through a half sibling. First, I add a set of dummy people and hypotheses to represent the full sibling line.
Note that all of the hypotheses were automatically renumbered when new ones were added. Don’t get too attached to the numbering at any one step of the process!
Then, I add another dummy child to Jasper and select “Define Half Relationships”.
Then, I can use the tick boxes to specify which of Jasper’s children are half siblings to this new addition, then click CLOSE.
Here is my finished hypothesis diagram:
Feel free to try this example for yourself and to add additional hypotheses. For example, try Doug as a child, grandchild, or great grandchild of Martha, Willard, and/or Anise to see what happens. I ended up with 18 hypotheses, like so:
Interpreting the Results
Using the last version of the tree, the first thing I notice is that some hypotheses (H1, H2, H4, H5, H7, H10, H13, H14, H15, and H16) have red flags with “Score = 0”. Those hypotheses are not possible given how much DNA Doug shares with his matches and what we currently know about the ranges for known relationships. I can “Remove Hypothesis” for those if I like (and the remaining hypotheses will be renumbered), or I can just ignore them.
The remaining hypotheses all have green flags and scores that are positive integers. Once the score = 0 hypotheses are ruled out, the remaining ones are assigned scores starting with 1 for the least likely (H3 in this case) and scaling up from there. From highest to lowest, the scores are: H6 (score = 293), H9 (288), H8 (204), H17 (201), H12 (8), H18 (4), H11 (2), and H3 (1).
What does this tell us? What it doesn’t tell us is precisely where Doug fits into the tree. But it does give us some guidance for where to look next. Four hypotheses have scores in the 200s, while the other viable hypotheses are all in single digits. Those low-score hypotheses are not where I’d focus my attention, efforts in contacting family, and testing dollars.
If money were tight, I’d focus solely on Jasper Pickle’s line, because that’s where three of the four highest ranked hypotheses are. If time were of the essence, I’d try to test descendants of Jasper, Maisie, Gerald, and James. With either approach, when the new results came in, I’d add them to the tree and re-evaluate the hypotheses.
But Wait! There’s More!
Scroll down below the tree, and you’ll see a listing of the hypotheses, their scores, and a status summary for each.
Scroll further for a “Collated Match Data” table, that summarizes the DNA matches, their relationships under each hypothesis, and the probability of each relationship given the cM amounts. (I couldn’t fit all 18 hypotheses into the screenshot.)
This table can give you insights into which matches are ruling out which hypotheses and sometimes can point to problems in your hypothesis tree. For example, if a certain hypothesis has high probabilities for all of the matches except one, which has zero probability, it’s worth a check to make sure that the cM amounts and relationships (especially full vs half) were entered correctly.
Finally, down at the bottom of the page are links to the main testing companies, should you need to buy additional DNA tests to confirm your hypotheses. Using these links won’t cost you anything extra and will help to subsidize the development of new tools for genealogy.
Housekeeping
WATO has some housekeeping features that let you save, share, and delete trees and switch among your saved trees.
With an account at DNA Painter, you can save multiple trees and switch among them at will. The most recent tree will be stored in your browser memory, and saved trees are accessible in the “Switch tree” pulldown.
For More Help
If you’re on Facebook, join the WATO group for support, strategies, and new developments.
Other posts in this series can be found here:
- Part 1 — Basic Probability
- Part 2 — Testing Hypotheses
- Part 3 — DNA Painter Look-up Tool
- Part 4 — GIs in Germany: Which Brother?
- Part 5 — Ruth: Using Probability to Guide Future Testing
- Part 6 — Ted, or When Close Relatives Aren’t Available
- Part 7 — The “What Are the Odds?” Tool (you’re here!)



















I have a similar tool that I’d love you to take a look at. It takes a more brute force approach, trying to fit the DNA test in at (what I hope is) all the positions in the tree.
You can find it at http://www.dnamatchanalyzer.com
Hi Leah – this is great!! Much easier the old table based method & aesthetically quite nice too! I was quickly able to build up a matrix of several hypotheses.
Fabulous! Glad to hear that you’re getting up to speed quickly.
Hi Leah,
Will this tool work when the target is related to the DNA testers on both maternal and paternal sides? Example: Adam and Meghan are 3rd cousins. Adam’s mother is Meghan’s father 2nd cousin. We need to find Adam’s father who is a relative of Meghan’s mother. We suspect Adam is a 2C1R of Meghan’s. Also, Adam’s parents are predicted 4th cousins according to gedmatch.
It’s not designed for endogamy, so you’ll have to take the scores with a grain of salt. Hopefully, it can still guide you in the right direction.
Great blog and great tool! I can already think of several cases where I want to try it out. Thanks to both you and Jonny Perl!
Also, many many thanks to Dr Andrew Millard, who worked out the math behind the “scores”. Couldn’t have done it without him!
I love this tool! For grins, I started with hypothesizing where I myself might fit into a network of 4C, 4C1R and 5C folks (determined by both paper genealogy and targeted testing, plus some lucky random matches). And for my FTDNA matches, I included all segments 6 cM and greater. (My reasoning, rightly or wrongly, is because MyHeritage and Ancestry also use 6 cM segments). My matches all shared from 25 cM to 80 cM, and the most likely hypothesis is that I would be a granddaughter of “Elizabeth” (who in fact is my grandmother). The other cool thing about this tool is that I now have a graphic of descendants of my 4th great-grandparents who have tested and match with me, and, interestingly, all their children who have descendants alive today have at least one person tested and at least one of those tested matching me.
Thanks to everyone who worked on this cool tool!
For FTDNA, I would use 7 cM.
I agree that the tool makes a great visual, even if you’re not actively working hypotheses. And it’s so easy to share a link with relatives!
Thank you so much for all your work and this wonderful series of posts! Invaluable in working with DNA Painter’s tools and bridging the gap between different testing platforms and the lack of a chromosome browser on AncestryDNA.
You’re welcome! Most of the time, a chromosome browser isn’t necessary. We just haven’t had a good tool for analyzing match data until now.
Thanks for this amazing tool. It was such an obvious idea – use probability math to weight possible relationships. But like many “obvious” ideas, someone had see it.
My great-grandparents were each married twice, with children from both marriages, and extended families in several cities. There were a few family dramas that fractured the families. And apparently there was at least one non-parent event, a child given up for an informal adoption, and a uncle marrying his niece. Add to that, my ggf and his brothers came from a small village and already shared extra dna. (My cousin and I share 1200 cM – the upper edge.)
Anyhow, I set up a large WATO tree that I can save a copy to work with when I find new matches. WATO has been extremely helpful. You’ve managed to make complex mathmatical calculations easy, with a clean UI. Thanks!
I’m so glad you like it! Bear in mind that it’s not currently designed to work with endogamy or uncle–niece marriages. Feel free to join the Facebook group if you have questions about using it or suggestions for improvement. https://www.facebook.com/groups/WhatAretheOdds/
I ran WATO on my current question: Which of several siblings was my bio GGF. I can identify my GGM, so this has to be a son. I set up 4 hypotheses: 3 sons and 1 daughter as a control. Scores: son#1, 182; son#2, 2; son#3, 1; dau, 60. OK, son#1 is likely by GGF.
Then I redid the same analysis using my brother’s cM data. Scores: son#1, 1; son#2, 2; son#3, 19; dau, 568. OOPs! Do I make the best guess that Mary is my GGF?
Yes, he is my full brother!
If you’re certain that you’ve correctly identified your GGM, then you can rule the daughter (Mary?) out, regardless of her scores. You can put yourself and your brother into the same analysis with a process I call twinning. For each of the matches in the WATO tree, create a duplicate and add your brother’s cM numbers to that “twin”. That will trick WATO into doing the calculations from both tress (yours and his) in a single tree.
Is there a tool/process I can use for double-cousins? I have two brothers who married two sisters, and I’m trying to help my cousin-match find where he fits in either family, since he matches both sides. I tried WATO, but then read that it doesn’t work with double-cousins. Right?
No, WATO doesn’t have the underlying statistics for double cousins. If your cousin-match is directly descended from one of the double marriages, you can simply ignore the matches who are descended from the other double marriage. If your cousin is descended from one of the four individuals in the double marriages (but not from one of the marriages themselves), then he shouldn’t have double relationships to the others and WATO should work.
Hi – I tried the WATO tool to test out hypotheses as to who my maternal grandfather might be, using 5 matches at 130, 122, 31, 17 and 15 cM, but have now been advised that anything under 40 is not useable. The example on the site uses some lower matches, so do you think the above would give a viable result, or not?
You can use matches below 40 cM, but you’ll get better results whhen most of your matches are above that threshold. That’s because the probability data on which WATO is based only goes down to 40 cM.
Thanks for clarifying that. It might be useful to add something to that effect to the instructions, as other people might also be in my position – ie we don’t have that many matches over 40cM to play with, so need to be aware of the limitations of the results.
There’s a FAQ in production that should be out soon.
I’m working a case now that is I think exceeding the limits of the WATO tool, for two reasons: (1) endogamy (2) he is related to descendants of two unrelated men, whose trees come together in a marriage, but there is no single ancestor for all of his matches.
The goal is to identify the birth father of the subject (an adoptee who has already identified his birth mother). He took the Ancestry DNA test and has HUNDREDS of high cM matches who all trace their ancestry back to two villages outside of Palermo, Sicily. Seems like most of them are related to each other with the same four or five surnames repeating over and over in their trees. So – endogamy.
Would it be OK to link to the two trees here, with only first names?
Since comments are moderated, here are the links – if it’s OK to post, you’ll approve the comment, otherwise no harm no foul!
https://dnapainter.com/tools/probability/view/429f600679f089ef
https://dnapainter.com/tools/probability/view/cdab622c47fa1bdb
These two trees meet in the marriage of Dominic 1877 and Antonina 1886. My working theory is that one of their sons, Joseph or John, is the subject’s birth father. I think the endogamy is skewing that – it thinks it’s most likely, in one of the charts, that the subject is the grandson of a man born in 1783!
WATO is not currently able to handle endogamy. Your best bet is to work the case “old school” by cross-referencing DNA matches, geographic locations, and targeted testing.
I’d like to see if I can fit a DNA match into my existing tree, but I don’t know how to get the cM shared from my target person to our common matches. Other than GEDMatch, which she is not on, is there another way to determine the cM shared from her rather than from me? using Ancestry’s match lists?
This post might help: https://thednageek.com/quick-tip-shared-matches-at-ancestrydna/
Hi, I’ve just started using WATO and I love the idea. But one thing bothers me, which I’ll illustrate with a real example.
My grandmorher was adopted. Using DNA I have found her birth mother (who was named on her birth certificate) but I haven’t yet been able to find her father (who wasn’t named). I have now tested the WATO tool on the matches that led me to identify her mother, and sure enough, the hypothesis that matches where I have found her was far and away the best. But as far as I can see, I could equally have thought I had identified her father – after all, the same tree applies in either case.
This is a particular issue for me because I have two groups of matches on different sides of the WATO tree. For a while I thought one group indicated her mother and one her father (i.e. her mother’s parents were cousins or second cousins). I decided that wasn’t correct, but it seems to me that WATO doesn’t give me information to judge that.
So, am I right in thinking that WATO doesn’t distinguish which parent the matches could relate to, but we have to know that from some other source? Thanks.
PS This doesn’t take away from how fantastic this tool is, but it will help me to know its limits.
Correct. By itself, WATO can’t tell whether you’re working the maternal side or the paternal one. You have to figure that out using other information.